PLANTEOME DEEP SEGMENT ANALYSIS MODULE¶
This module segments a marked object (creating a graphical object) within an input image.
Then the module will classify either the entire original image or the segment created in the first step. This uses PyTorch in order to do this deep segmentation.
Reference¶
- Module Development Guide: https://github.com/pndaly/BisQue_Platform_Guide
- Sample Deep Learning Module: Planteome Deep Segment Analysis
Pre-requisite: link¶
- Working bisque environment at http://loup.ece.ucsb.edu:8088/
- Docker should be enabled and setup on this environment
- The bisque deployment server should have access to good CPU & RAM since Torch & Scikit-image is used
- Access to module folder for deploying this module (Say at ~/bisque/module)
Add/Deploy the Planteome module¶
- Identify the bisque module folder at path ~/bisque/module
- Login to http://loup.ece.ucsb.edu:8088/ with credentials admin:admin
- Download the Planteome module to the bisque module folder
- Register the module to Bisque by opening the Module Manager
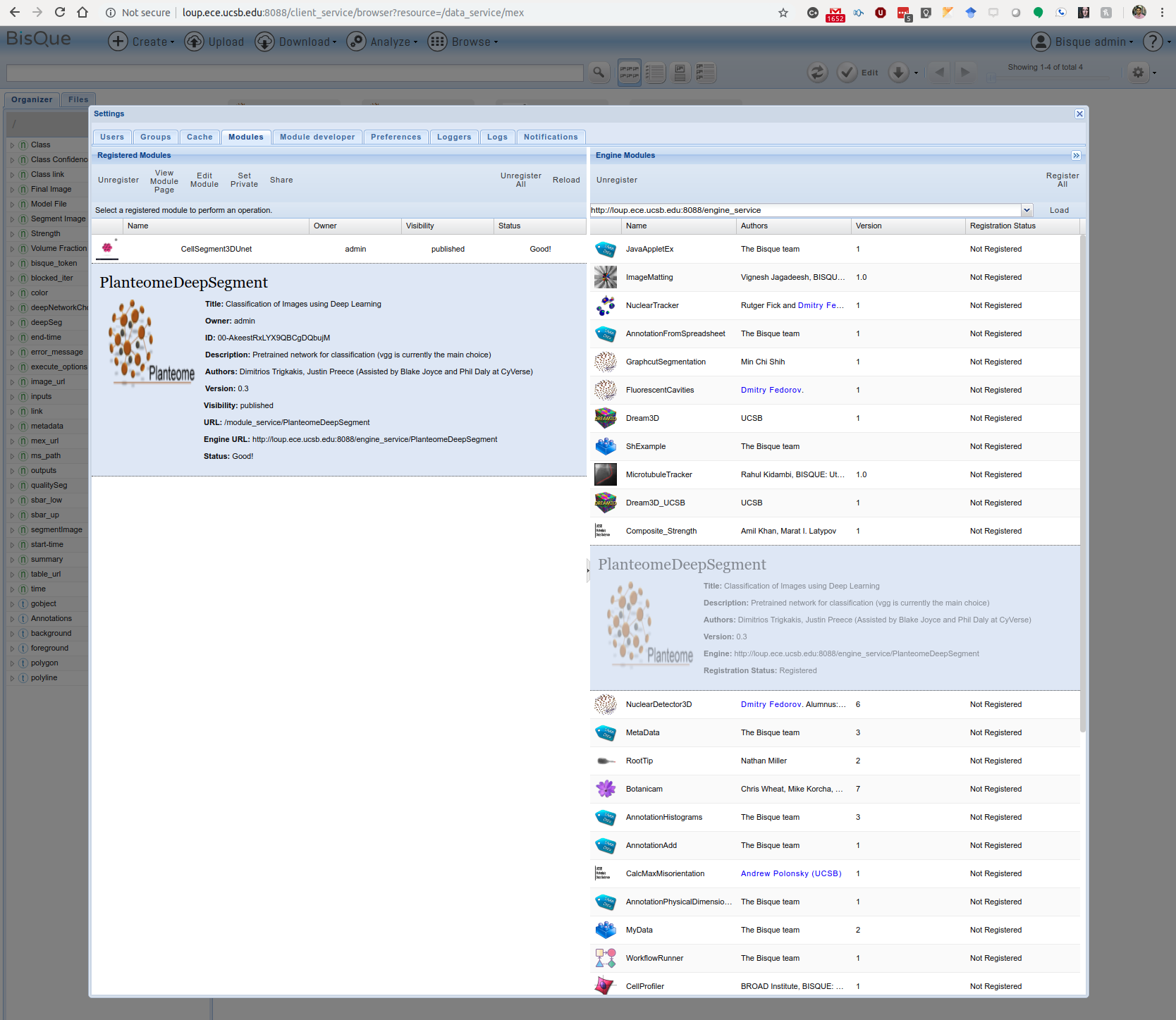
- Provide the engine URL(http://loup.ece.ucsb.edu:8088/engine_service) in the Engine service section and click load. This will list the modules by querying the engine service.
- Select the module named "Planteome", drag this module to the left pane and drop it to register.
Build/Configure the module¶
- Edit the runtime-module.cfg with relevant configuration
- Changed the docker.hub configuration to biodev.ece.ucsb.edu:5000
- View/Edit the Dockerfile for the relevant packages
- Edited versions for numpy\==1.16.1 & scikit-image\==0.14.2
- Source to your bisque python environment "workon bqenv" for installation
- pip install -r requirements.txt
- python setup.py
This will install all the dependency in the modules requirements.txt file, build modules code, and create a docker image for running it. Based on your "docker.hub" configuration the setup will push the docker image to registry as [biodev.ece.ucsb.edu:5000/bisque_uplanteome]
Steps to run the algorithm from Bisque Client¶
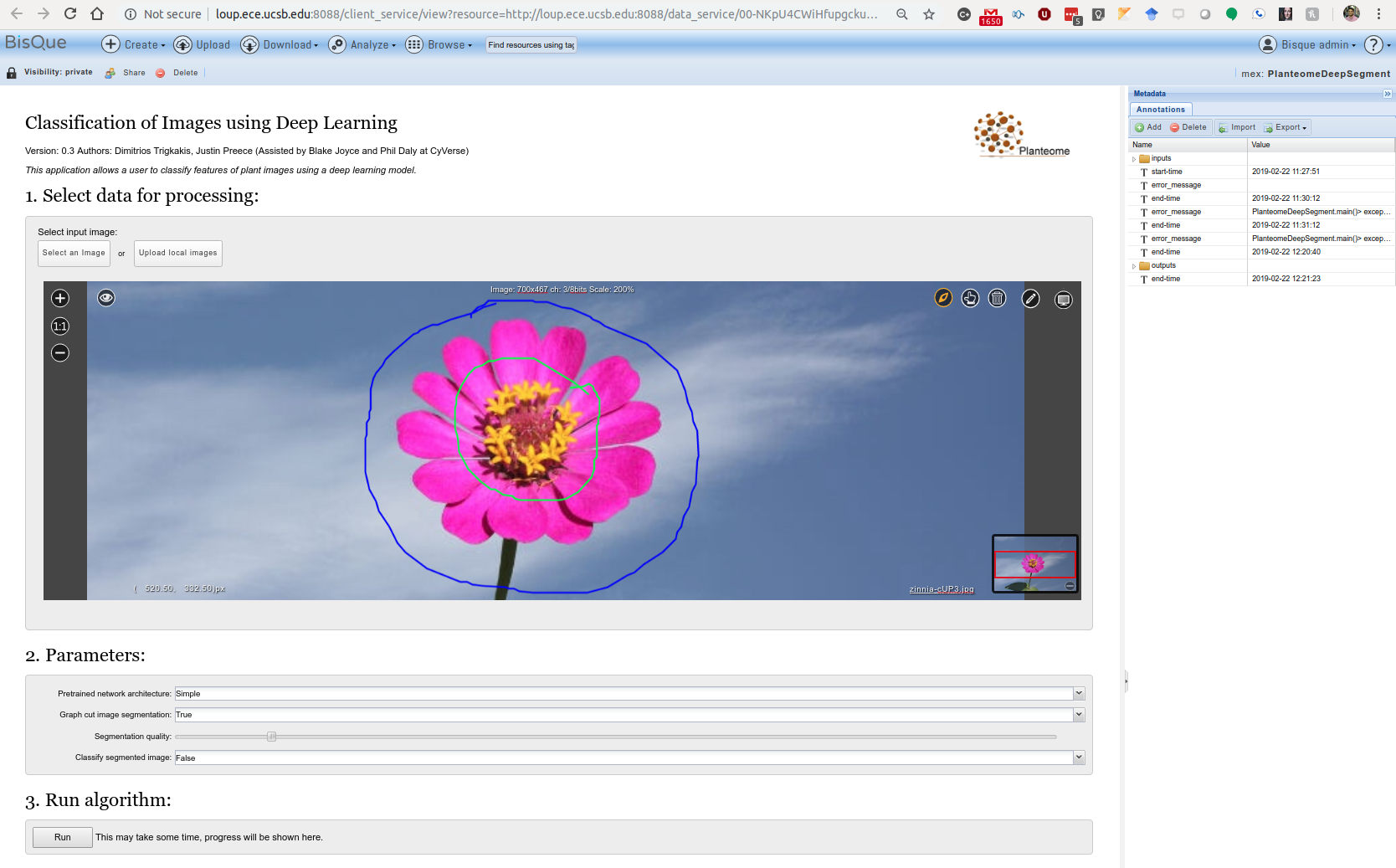
- Select an image to be analyzed.
-
The foreground/background annotation tooltip can be found on the top-right of the module image viewer. Mark the part of the image to be segmented with foreground line(s) annotation(s). Mark the image with background annotations around the object to be segmented.
-
Select which deep classifier to use, whether to segment the image, the segmentation quality, and whether to classify the entire image or the segmented object instead.
-
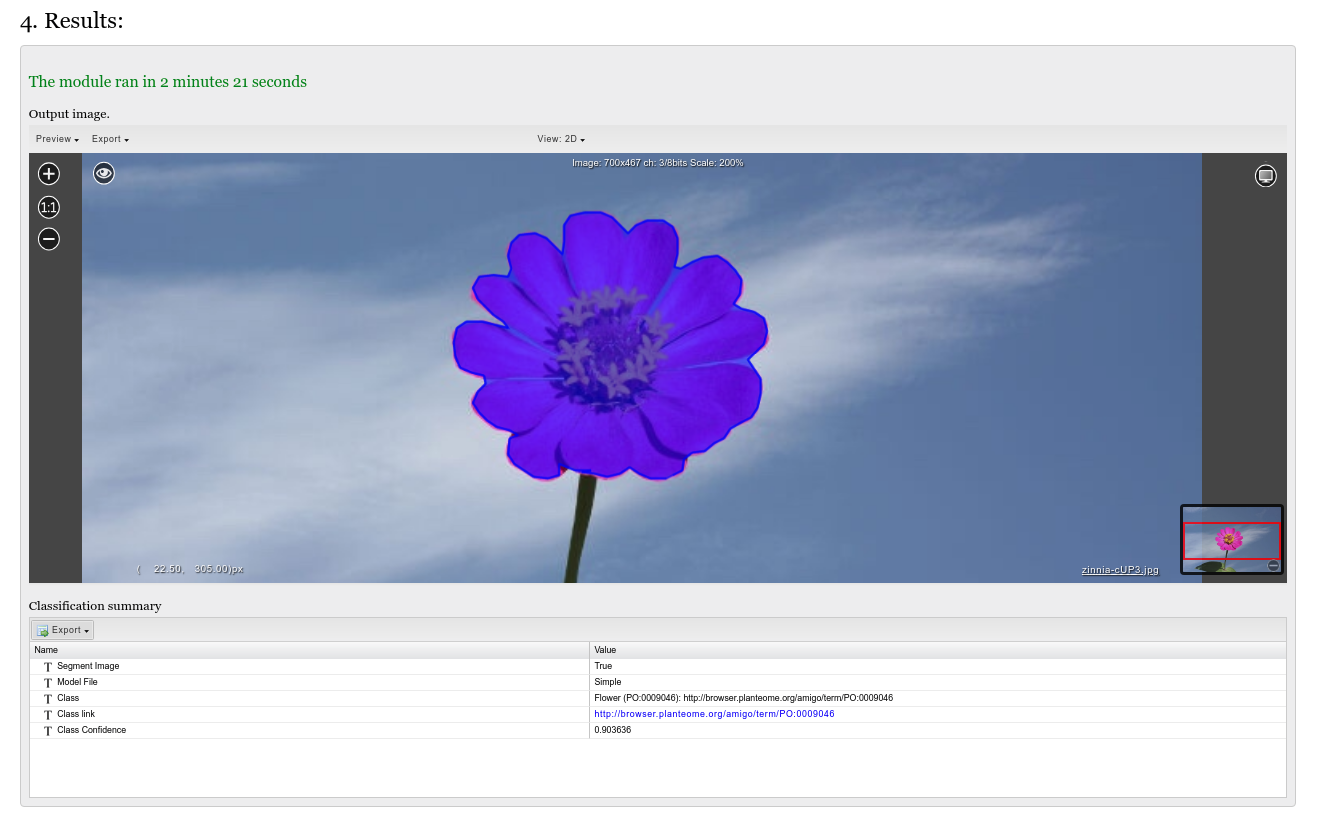
Press the 'RUN' button. Analysis may take some time depending on the image and segmentation quality.
-
Results are given in visual and table formats, depending on whether the segmentation and classification functionalities respectively were enabled in the options.
Isolated/Development test setup¶
Make sure you have annotated the image and have an a mex identifier availablefor manual test/run. This can be done by, - Opening the module and select image - Annotate the image as per the directions above - Configure and run the module once - Make note of the mex URL for this run by looking at the docker_run.log in the staging folder. This will be used for replaying the test run from the modules folder.
Additional module execution information - When we annotate the image and click RUN on the module user interface - The Planteome module is created from biodev.ece.ucsb.edu:5000/bisque_uplanteome:latest
``` docker create biodev.ece.ucsb.edu:5000/bisque_uplanteome \ python PlanteomeDeepSegment.py \ http://loup.ece.ucsb.edu:8088/module_service/mex/00-NKpU4CWiHfupgckuXBNFDd \ admin:00-NKpU4CWiHfupgckuXBNFDd Simple True 3 False ```
- This module is run with the hash id of the docker create
Setup/Run the docker container for test¶
- Build the docker container
- docker build --no-cache -t bisque_uplanteome -f Dockerfile .
- docker build -t biodev.ece.ucsb.edu:5000/bisque_uplanteome .
- Run the container and bash into it
- docker run -it --net=host biodev.ece.ucsb.edu:5000/bisque_uplanteome bash
- test run the code on mex identifier
- python PlanteomeDeepSegment.py http://loup.ece.ucsb.edu:8088/module_service/mex/00-NKpU4CWiHfupgckuXBNFDd admin:00-NKpU4CWiHfupgckuXBNFDd Simple True 3 False
Version: 0.3 Author(s): Dimitrios Trigkakis, Justin Preece